|
Biogeme: Python Library
2.5
|
|
Biogeme: Python Library
2.5
|
Class representing the product of the same expression applied to a list of data. More...
Public Member Functions | |
| def | __init__ (self, term, iteratorName, positive=False) |
| def | getExpression (self) |
 Public Member Functions inherited from bio_expression.Expression Public Member Functions inherited from bio_expression.Expression | |
| def | __init__ (self) |
| Constructor. | |
| def | getExpression (self) |
| def | getID (self) |
| def | __str__ (self) |
| def | __neg__ (self) |
| def | __add__ (self, expression) |
| def | __radd__ (self, expression) |
| def | __sub__ (self, expression) |
| def | __rsub__ (self, expression) |
| def | __mul__ (self, expression) |
| def | __rmul__ (self, expression) |
| def | __div__ (self, expression) |
| def | __rdiv__ (self, expression) |
| def | __truediv__ (self, expression) |
| Support for Python version 3.x. More... | |
| def | __rtruediv__ (self, expression) |
| Support for Python version 3.x. More... | |
| def | __mod__ (self, expression) |
| def | __pow__ (self, expression) |
| def | __rpow__ (self, expression) |
| def | __and__ (self, expression) |
| def | __or__ (self, expression) |
| def | __eq__ (self, expression) |
| def | __ne__ (self, expression) |
| def | __le__ (self, expression) |
| def | __ge__ (self, expression) |
| def | __lt__ (self, expression) |
| def | __gt__ (self, expression) |
Public Attributes | |
| function | |
| iteratorName | |
| positive | |
| operatorIndex | |
 Public Attributes inherited from bio_expression.Expression Public Attributes inherited from bio_expression.Expression | |
| operatorIndex | |
Class representing the product of the same expression applied to a list of data.
The concept of iterators identifies a sequence such that, for each instance, the value of the variables is read from the data file, and an expression can be evaluated. The two expressions described in this section consider one iterator and one expression, and evaluate the expression for each instance defined by the iterator. A product can then be computed. The following example computes the loglikelihood for a model with panel data.
The iterator personIter iterates on each individual in the file, characterized by the identifier Id. The iterator panelObsIter iterates on the observations (that is, the rows in the data file) associated with the current individual.
Assuming that prob is the likelihood of the observation in one raw, for a given set of draws, the following quantities are computed:
![\[ \mbox{condProbIndiv} = P(y_1,\ldots,y_T|\xi_n) = \prod_t P(y_t|\xi_n)\]](form_9.png)
![\[ \mbox{probIndiv} = \int_{\xi_n}P(y_1,\ldots,y_T|\xi_n) \approx \sum_r P(y_1,\ldots,y_T|\xi_r) / R \]](form_10.png)
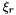 are the R draws generated from
are the R draws generated from 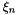 .
.
![\[ \mbox{loglikelihood} = \sum_n \log(\sum_r P(y_1,\ldots,y_T|\xi_r) / R) \]](form_13.png)
Definition at line 733 of file bio_expression.py.
| def bio_expression.Prod.__init__ | ( | self, | |
| term, | |||
| iteratorName, | |||
positive = False |
|||
| ) |
| term | any valid bio_expression |
| iteratorName | name of an iterator already defined |
| positive | Set it to True if all factors of the product are strictly positive. In that case, it will be computed as
|
Definition at line 739 of file bio_expression.py.